News
22.04.2024
Official Graduation of Brennen and Amer

At the graduation last Friday Brennen Heames and Amer Ghalawinji officially received their doctoral degrees. Brennen, who was part of our own group, came all the way back from the UK to receive his degree. He investigated the birth of de novo genes in Drosophila and looked more closely at their structural properties in comparison to random proteins. Amer from Prof. Monika Stoll's group looked at as part of the EvoPad graduate schoolthe effect of lncRNAs on genetic predisposition to heart failure with Erich as co-supervisor. Congratulations to both of them!
18.04.2024
Paper out in Genome Biology and Evolution!
Margaux Aubel, Filip Buchel, Brennen Heames, Aun Jones, Ondrej Honc, Erich Bornberg-Bauer, Klara Hlouchova Genome Biology and Evolution
Together with our collaborators Klara Hlouchova, Filip Buchel and Ondrej Honc in Prague, we looked into the folding propensities of de novo emerged proteins. Akin to random sequences, proteins that emerge de novo from noncoding DNA are usually predicted to be highly disordered as they inherit no structure from their ancestor, unlike proteins evolving through duplication. Most studies to-date have relied on computational prediction of the structural properties of de novo proteins. Here, we show experimentally that, while most of our putative human de novo proteins are highly disordered, some contain propensity for structure and globularity, seen most clearly in the oldest de novo proteins in our dataset. With that we aim to lay the groundwork for experimental verification of hypotheses regarding the structural evolution of de novo proteins.
20.01.2024
Paper out in Proteins!
Lasse Middendorf, Lars Eicholt Proteins: Structure, Function and Bioinformatics
The paper, "Random de novo and Conserved Proteins: How Structure and Disorder Predictors Perform Differently," by Lasse and Lars, investigates the structural characteristics of de novo, random, and conserved proteins using protein structure prediction tools like AlphaFold2 and ESMFold. It examines how these tools perform with such proteins lacking sequence identity and highlights the differences in structural predictions and confidence scores (pLDDT) between these protein. The study also explores the influence of sequence length and Multi-Sequence Alignment (MSA) depth on prediction outcomes. The findings suggest a unique structural and disorder profile for de novo and random proteins in which pLDDT correlates negatively with beta-sheets and positively with disorder and alpha-helices. In conserved proteins pLDDT normally correlates negatively with disorder and is often used as an indicator of disordered regions.
12.01.2024
Master Thesis Defense Of Alina and Lasse

Starting into the New Year with not only one, but two great Master Thesis defenses! First Alina Berger defended her thesis on the "Evolution of the CatSpermasome". She looked into the evolutionary history of the sperm-specific ion channel, which is essential for male fertility to track down its emergence throughout vertebrates and detected signatures of positive selection along the phylogenetic tree. Her second thesis advisor was Timo Strünker from the UKM.
Then Lasse Middendorf presented and successfully defeded his thesis on "
4.12.2023
PhD defence Brennen Heames

Last friday we had the honour to listen to Brennen's PhD defence about "Exploring the potential of de novo gene emergence to support protein-coding evolutionary novelty". It was a great talk and it has been a pleasure to have him in the group!
10.11.2023
Anna and Erich in Texas

Erich and Anna represented the group at the SMBE Sattelite Meeting on "De novo Gene Birth" in College Station, Texas. The meeting was beautifully organised by Claudio Casola, Li Zhao, Victor Luria and Nikolaos Vakirlis. We are looking forward to seeing you all again, hopefully at the next SMBE in Puerto Vallarta, Mexico next July.
24.10.2023
Biospektrum Article on de novo Proteins

The experimentalists of our group Margaux, Lars and Andreas wrote an article with Erich for the german magazine Biospektrum about our work on de novo proteins. In the article the focus is mainly on the experimental side of the research, which is explained to a broad audience not familar to de novo gene evolution.
30.07.2023
SMBE in Italy

The SMBE 2023 took place in the beautiful Ferrara in Italy. Erich and Claudia Alvarez-Carreño organised the symposium on "Novel proteins and their emergence from LUCA until today" where Lars gave his talk. Elias presented his work in the Symposium on "Computational evolutionary genomics in the era of machine learning" and Margaux as part of the "Graduate Students Excellence Awards" Symposium. Bharat presented his poster in "Evolutionary biology through a functional genomics lens". We hope to see many people again next year in Mexico or at the SMBE Sattelite Meeting on "De novo Gene Birth" in Texas in November!
25.07.2023
Population genomics reveals mechanisms and dynamics of de novo expressed open reading frame emergence in Drosophila melanogaster
Anna Grandchamp, Lucas Kühl, Marie Lebherz, Kathrin Brüggemann, John Parsch, Erich Bornberg-Bauer Genome Research

Congratulations Anna and Marie on this really cool new paper! They found that novel genes are essential for evolutionary innovations and that they differ substantially even between closely related species. Some of those novel genes arise de novo, that is, from previously noncoding DNA. To characterize the underlying mutations that allowed de novo gene emergence and their order of occurrence, homologous regions must be detected within noncoding sequences in closely related sister genomes. Here they sequenced and assembled genomes with long-read technology and the corresponding transcriptomes from inbred lines of Drosophila melanogaster, derived from seven geographically diverse populations (see picture on the left). The new data suggests a rapid turnover of novel transcripts and more frequent gain and loss of transcription than the creation of ORFs. The highly mutable genomic regions around TEs provide new features that enable gene birth. In conclusion, de novo ORFs have a high birth-death rate, are rapidly purged, but surviving de novo ORFs spread neutrally through populations and within genomes.
13.07.2023
Protein Evolution Meeting Münster

We had a great Protein Evolution Meeting in Münster (PEMM) this week! Thanks to all speakers, visitors, poster creators and helpers. We hope to see everyone again soon and look forward to the next meeting of this kind.
10.06.2023
University News

Great article about our group and our research on de novo proteins from our university newspaper! Check it out here.
18.04.2023
Neutral models of de novo gene emergence suggest that gene evolution has a preferred trajectory
Bharat Ravi, Erich Bornberg-Bauer Molecular Biology and Evolution

Cheers and congratulations to Bharat! His new paper is about the theory behind the emergence of de novo genes.
New protein coding genes can emerge from genomic regions that previously did not contain any genes, via a process called de novo gene emergence. To synthesize a protein, DNA must be transcribed as well as translated. Both processes need certain DNA sequence features. Stable transcription requires promoters and a polyadenylation signal, while translation requires at least an open reading frame (ORF). We develop mathematical models based on mutation probabilities, and the assumption of neutral evolution, to find out how quickly genes emerge and are lost. We also investigate the effect of the order by which DNA features evolve, and if sequence composition is biased by mutation rate. We rationalize how genes are lost much more rapidly than they emerge, and how they preferentially arise in regions that are already transcribed. Our study not only answers some fundamental questions on the topic of de novo emergence but also provides a modeling framework for future studies.
07.04.2023
Experimental characterization of de novo proteins and their unevolved random-sequence counterparts
Brennen Heames, Filip Buchel, Margaux Aubel, Vyacheslav Tretyachenko, Dmitry Loginov, Petr Novák, Andreas Lange, Erich Bornberg-Bauer, Klara Hlouchova Nature Ecology & Evolution

Together with the KHlab in Prague we published our study on de novo proteins compared to random proteins. De novo gene emergence provides a route for new proteins to be formed from previously non-coding DNA. Proteins born in this way are considered random sequences and typically assumed to lack defined structure. Taking putative de novo proteins identified in human and fly, we experimentally characterize a library of these sequences to assess their solubility and structure propensity. We compare this library to a set of synthetic random proteins with no evolutionary history. Bioinformatic prediction suggests that de novo proteins may have remarkably similar distributions of biophysical properties to unevolved random sequences of a given length and amino acid composition. However, upon expression in vitro, de novo proteins exhibit moderately higher solubility which is further induced by the DnaK chaperone system. We suggest that while synthetic random sequences are a useful proxy for de novo proteins in terms of structure propensity, de novo proteins may be better integrated in the cellular system than random expectation, given their higher solubility.
06.04.2023
HFSP Awards 2023

Happy to announce that the lab just received its 4th HFSP grant! “New Kids on the Block: how De Novo emerged micropeptides rewire cellular networks”. This time with Anne-Ruxandra Carvunis and Christine Brun. HFSP Awards 2023
21.03.2023
Münster Evolution Meeting

Last week our group participated in the second Münster Evolution Meeting (MEM). There were many interesting talks and great posters and we were lucky to meet so many great people. It was a nice conference and many of us had the opportunity to present their research in a talk (like Anna Grandchamp here in the picture) or during the poster sessions. We want to thank the organising team from the Gadau group again and hope to see everyone again at the next MEM!
28.02.2023
Guest Researcher Aleksandra Marsavelski

For one exciting month we have a guest researcher from Zagreb, Croatia in our group. Aleksandra Marsavelski is visiting us as a WiRe fellow to look at plastic degrading enzymes. Together with our group she wants to reconstruct ancestors of enzymes with a known plastic degrading activity and enhance their function. This can help us in the future to recycle plastics in an environmentally friendly way. For more information and news, check out the WiRe website of the University here.
03.02.2023
Introducing creative destruction as a mechanism in protein evolution
Margaux Aubel, Erich Bornberg-Bauer Proceedings of the National Academy of Sciences

In this commentary we discuss the recent paper from Alvarez-Carreño et al. about the evolution of new folds. In particular, we look at ancient folds from the dawn of the RNA-protein world and how new and still existing folds may have originated through reshaping of the old folds. The mechanism introduced by Alvarez-Carreño et al., called "creative Destruction" describes the process of innovation through the “destruction” of an old product and thereby generating a new one. An analogy from everyday life is the development of an mp3 player from a cassette player. The old cassette player, lost its overall form and the need of a cassette tape to develop into an mp3 player. Form and function of both products are similar while some aspects of the ancestral product have been ‘destroyed’, i.e. cassette, bulky structure and loudspeaker, to create the new features, i.e. memory space, small, headphones.
01.02.2023
Blattodea Conference in April

27.01.2023
Kai's Master Thesis handed in and defended

Our Master student Kai has succesfully defended his Thesis last December on "Structural analysis of putative de novo protein Atlas and its ancestors". After mastering many obstacles in the wet lab, Kai found his true calling during the computational analyses. Now he is heading off to new computational adventures and we wish him all the best of luck on his journey!
07.12.2022
IEB seminar by Birte Höcker

Birte Höcker from the University of Bayreuth gave an IEB seminar on the Evolution and Design of Protein Folds. Thanks for visiting our group and the IEB!
01.12.2022
Life? Symposium in Hanover - Ghost in the protein

We had a great symposium at the Xplanatorium in Hanover Herrenhausen from the 29th to 30th of November. Together with our collaborators Ylva Ivarsson from Uppsala, Florian Hollfelder from Cambridge and Klara Hlouchova from Prague we look into the early evolution of proteins from random sequence space. Our project on the emergence of new proteins by de novo mechanism is funded by the Volkswagen Stiftung Life? initiative together with several other interesting projects.
30.11.2022
2nd Münster Evolution Meeting from 13th-16th March 2023. Registration is open!

We are happy to announce that MEM is finally back! See you all in Münster from 13th -16th March 2023. The Münster Evolution Meeting (MEM) aims to provide a forum for all Evolutionary Biologists working across different fields. Besides having the opportunity to share and learn about excellent research in evolutionary biology MEM also aims at bringing together Evolutionary Biologists working in German-speaking countries in a smaller setting, to allow for intensive networking and discussion. Münster is a welcoming and vibrant university town, offering a perfect venue. It was the home of Prof. Dr. Bernhard Rensch, who contributed significantly to the “Modern Evolutionary Synthesis”. Click here to register!
03.11.2022
Eusocial Transition in Blattodea: Transposable Elements and Shifts of Gene Expression
Juliette Berger, Frédéric Legendre, Kevin-Markus Zelosko , Mark C. Harrison, Philippe Grandcolas, Erich Bornberg-Bauer, Bertrand Fouks Genes

Using available genome and transcriptome of cockroaches and termites as well as de novo transposon annotations, our study brings further evidence on the role of transposon-aided adaptations during termite eusocial transition. Our study reveals that transposon insertions within differentially expressed genes (DEGs) among castes in termites and life-stages (corresponding to termite castes) in cockroaches display opposite patterns. Furthermore, while in cockroaches transposon families do not differ in their insertion frequency within DEGs, retrotransposon and DNA transposon insertions are more frequent in worker- and queen-biased genes, respectively. In addition, the insertion of transposons in queen-biased genes in termites are more frequent in genes involved in reproduction, ageing, and immunity, functions essential for queen phenotype; while transposon insertion in termite worker-biased genes are more frequent in genes involved in the regulation of gene expression and behaviour, functions associated with phenotypic plasticity and the apparition of a sterile caste. In conclusion, our study highlights the role of transposon during the eusocial transition in Blattodea, potentially through shifts in gene expression.
12.10.2022
Sarah's Master Thesis Celebration & PhD start

After Sarah's successful Master thesis defence we had some delicious Flammkuchen together with her family. The title of her thesis was "The role of selection in the emergence of caste-biased genes in termites" and she was under the amazing supervision of Mark. We are very happy that she now started her PhD in the group on the evolutionary genomics of sociality in beetles funded by GEvol (SPP 2349)
11.10.2022
Domain Evolution of Vertebrate Blood Coagulation Cascade Proteins
Abdulbaki Coban, Erich Bornberg-Bauer, Carsten Kemena Jornal of Molecular Evolution

Here we examined proteomes of 21 chordates, of which 18 are vertebrates, to reveal the modular evolution of the blood coagulation cascade. Within the vertebrate coagulation protein set, almost half of the studied proteins are shared with jawless vertebrates. Domain similarity analyses revealed that there are multiple possible evolutionary trajectories for each coagulation protein. During the evolution of higher vertebrate clades, gene and genome duplications led to the formation of other coagulation cascade proteins.
04.10.2022
Group retreat in Pfaffenhofen a. d. Roth

We just got back from our awesome second group retreat this year! A lot of great presentations and talks, especially from and with our special guest Richard Goldstein. Apart from the serious scientific topics, we went for a beautiful walk to see the famous "Küssende Sau" (kissing pig). Stay tuned for more pictures...
29.09.2022
Priority Programme “Genomic Basis of Evolutionary Innovations (GEvol)” (SPP 2349) kick-off meeting in Münster
Official website of the SPP GEvol

The kick-off meeting for the DFG Priority Programme “Genomic Basis of Evolutionary Innovations (GEvol)” (SPP 2349) coordinated by Erich was the first in-person meeting of nearly all of the 40 involved PIs! The goal of the Priority Programme is to exploit new methods to reveal in the insect taxon the role of: coding vs. regulatory changes, transposable elements, epigenetic regulation, gene family evolution, copy number dynamics, structural genomic rearrangements etc. in trait evolution by using multiple cutting edge quantitative OMICs resources (e.g. genomics, transcriptomics and epigenomics). Eventually, the emerging hypotheses shall be tested by functional genetics experiments where possible.
13.09.2022
Vector redesign and in-droplet cell-growth improves enrichment and recovery in live Escherichia coli
Bernard D. G. Eenink, Tomasz S. Kaminski, Erich Bornberg-Bauer, Joachim Jose, Florian Hollfelder, Bert van Loo Microbial Biotechnology

In this article we compare different methods to sort large libraries of enzymes presented on the surface of E. coli, and recovering the intact cells without lysis. We changed a protocol in which single cells encapsulated in a microdroplet are measured using Fluorescence-activated droplet sorting (FADS) to a protocol were cells are grown inside a microdroplet after encapsulation of a single cell, resulting in multiple copies of the same variant inside each droplet. We find that growing cells inside a microdroplet prior to FADS increases both the amount of cells recovered as well as reducing the amount of false positives.
30.08.2022
The role of transposable elemets in the evolution of insect socal complexity - Erich Bornberg-Bauer at the HFSP meeting in Paris

This time the group leader Erich Bornberg-Bauer himself presented our work on the role of transposable elements in termites. The 21st HFSP Awardees meeting took place in beautiful Paris this year. The poster is based on work by postdoc Mark Harrison and former postdoc Evelien Jongepier from our group in cooperation with Mireille Vasseur-Cognet (Paris), Hei Sook Sul (Berkeley) and Wilhelm de Beer (Pretoria).
We compared whole-genome levels of repeat element transcription in the fat body of female workers, kings, and five different
queen stages. Those queens can live over 20 years, maintaining near maximum reproductive output! The sterile workers
only live weeks or months. We found substantially higher TE activity in workers than in
reproductives. Furthermore, relative TE expression decreased with age in queens, due
to a significant upregulation of the PIWI-pathway in 20-year-old queens. Our results
suggest a caste- and age-specific regulation of the PIWI-pathway has evolved in higher
termites that is analogous to germ-line-specific activity in individual organisms,
promoting reproductive fitness even at high age.
30.08.2022
Evidence for a conserved queen-worker genetic toolkit across slave-making ants and their ant hosts
Barbara Feldmeyer, Claudia Gstöttl, Jennifer Wallner, Evelien Jongepier, Alice Séguret, Donato A. Grasso, Erich Bornberg-Bauer, Susanne Foitzik, Jurgen Heinze Molecular Ecology

The ecological success of social Hymenoptera (ants, bees, wasps) depends on the division of labour between the queen and workers. Each caste exhibits highly specialized morphology, behaviour, and life-history traits, such as lifespan and fecundity. Despite strong defences against alien intruders, insect societies are vulnerable to social parasites, such as workerless inquilines or slave-making ants. Here, we investigate whether gene expression varies in parallel ways between lifestyles (slave-making versus host ants) across five independent origins of ant slavery in the “Formicoxenus-group” of the ant tribe Crematogastrini. As caste differences are often less pronounced in slave-making ants than in nonparasitic ants, we also compare caste-specific gene expression patterns between lifestyles. We demonstrate a substantial overlap in expression differences between queens and workers across taxa, irrespective of lifestyle. Caste affects the transcriptomes much more profoundly than lifestyle, as indicated by 37 times more genes being linked to caste than to lifestyle and by multiple caste-associated modules of coexpressed genes with strong connectivity. However, several genes and one gene module are linked to slave-making across the independent origins of this parasitic lifestyle, pointing to some evolutionary convergence. Finally, we do not find evidence for an interaction between caste and lifestyle, indicating that caste differences in gene expression remain consistent even when species switch to a parasitic lifestyle. Our findings strongly support the existence of a core set of genes whose expression is linked to the queen and worker caste in this ant taxon, as proposed by the “genetic toolkit” hypothesis.
23.08.2022
Anna Grandchamp and Alina Mikhailova present their work at ESEB in Prague

Our postdoc Anna Grandchamp presented her poster on proto-gene emergence and PhD student Alina Mikhailova gave a talk about domain rearrangements in social termites. Awesome work by both of them!
21.07.2022
MC Harrison on ICE (International Congress of Entomology)

Our postdoc Mark C Harrison presented his work on social insects this week on the International Congress of Entomology (ICE) in Helsinki, Finnland. Check out his latest publications in Open Biology and Genome Biology and Evolution
21.07.2022
Complex regulatory role of DNA methylation in caste- and age-specific expression of a termite
Mark C. Harrison, Elias Dohmen, Simon George, David Sillam-Dussès, Sarah Séité, Mireille Vasseur-Cognet Open Biology

The reproductive castes of eusocial insects are often characterized by extreme lifespans and reproductive output, indicating an absence of the fecundity/ longevity trade-off. The role of DNA methylation in the regulation of caste- and age-specific gene expression in eusocial insects is controversial. While some studies find a clear link to caste formation in honeybees and ants, others find no correlation when replication is increased across independent colonies. Although recent studies have identified transcription patterns involved in the maintenance of high reproduction throughout the long lives of queens, the role of DNA methylation in the regulation of these genes is unknown. We carried out a comparative analysis of DNA methylation in the regulation of caste-specific transcription and its importance for the regulation of fertility and longevity in queens of the higher termite Macrotermes natalensis. We found evidence for significant, well-regulated changes in DNA methyl- ation in mature compared to young queens, especially in several genes related to ageing and fecundity in mature queens. We also found a strong link between methylation and caste-specific alternative splicing. This study reveals a complex regulatory role of fat body DNA methylation both in the div- ision of labour in termites, and during the reproductive maturation of queens.
13.07.2022
Heterologous expression of naturally evolved putative de novo proteins with chaperones
Lars A. Eicholt, Margaux Aubel, Katrin Berk, Erich Bornberg-Bauer, Andreas Lange Protein Science

Today, we know that proteins do not only evolve by duplication and divergence of existing proteins but also arise from previously non-coding DNA. These proteins are called de novo proteins. Their properties are still poorly understood and their experimental analysis faces major obstacles. Here, we aim to present a starting point for soluble expression of de novo proteins with the help of chaperones and thereby enable further characterization.
Over the past decade, evidence has accumulated that new protein-coding genes can emerge de novo from previously non-coding DNA. Most studies have focused on large scale computational predictions of de novo protein-coding genes across a wide range of organisms. In contrast, experimental data concerning the folding and function of de novo proteins are scarce. This might be due to difficulties in handling de novo proteins in vitro, as most are short and predicted to be disordered. Here, we propose a guideline for the effective expression of eukaryotic de novo proteins in Escherichia coli. We used 11 sequences from Drosophila melanogaster and 10 from Homo sapiens for heterologous expression. The candidate de novo proteins have varying secondary structure and disorder content. Using multiple combinations of purification tags, E. coli expression strains, and chaperone systems, we were able to increase the number of solubly expressed putative de novo proteins from 30% to 62%. We found that, overall, proteins with higher predicted disorder were easier to express.
26.04.2022
Janina (GadauLab) and Lars elected as IEB student representatives

Our PhD student Lars and Janina Rinke from the lab of Jürgen Gadau were elected as IEB student representatives for the next two years!
26.04.2022
Master Defence Christopher Finke: "Genome annotation and comparison of genomic features between slave-maker ants and their hosts"

Our dear student Christopher Finke has successfully defended his Master thesis yesterday evening. Supervised by Alice Séguret he looked into differences between slave-maker ants and their host species and was able to generate valuable genome annotations. Christopher started out as a Bachelor student in the lab with us and moved on to computational work during his Master studies. Congratulations and good luck in Berlin!
21.04.2022
Plant biodiversity assessment through soil eDNA reflects temporal and local diversity
María Ariza, Bertrand Fouks, Quentin Mauvisseau, Rune Halvorsen, Inger Greve Alsos, Hugo de Boer Methods in Ecology and Evolution

The use of environmental DNA as a proxy to identify species has increased exponentially in the last few years. However, how good eDNA relates with actual species presence/abundance remains elusive. Our study fill this gap, comparing both soil eDNA and visual assessments of plants in Norway. It shows that soil eDNA allows to describe well local plant biodiversity, with an average 60% match with visual survey for vascular plants, and also to recover past vegetation biodiversity. Our soil eDNA method then provide a great tool to assess rapidly plant biodiversity in any seasons.
08.04.2022
PhD students Margaux and Lars presenting their posters at Apfed22 in Bayreuth

Our PhD students Margaux Aubel and Lars Eicholt presented their most recent work on soluble expression of putative de novo proteins at Apfed22 in Bayreuth! If you did not attend, please check out their preprints here and here. We would like to thank all the organizers for this amazing hybrid conference. The speaker line-up was diverse both in backgrounds and research areas, while reaching gender parity!
22.02.2022
Co-expression analyses, combined with lipidomics and metabolomics, uncover lifespan prolonging mechanisms in extremely long-lived and highly fertile termite queens
Sarah Séité, Mark C Harrison, David Sillam-Dussès, Roland Lupoli, Tom J M Van Dooren, Alain Robert, Laure-Anne Poissonnier, Arnaud Lemainque, David Renault, Sébastien Acket, Muriel Andrieu, José Viscarra, Hei Sook Sul, Z Wilhelm de Beer, Erich Bornberg-Bauer , Mireille Vasseur-Cognet Communications Biology
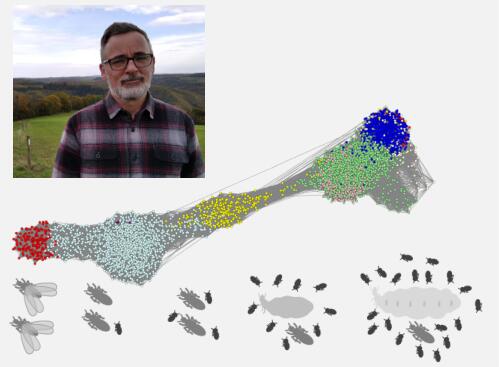
Some insects are eusocial, which means that their society is organized into different castes which carry out specific colony tasks. In termites, for example, the “king” and the “queen” are involved in reproduction, while the “workers” are involved in resource gathering and brood-care. Macrotermes natalensis is a species of higher termite that creates large complex colonies, in which the king and the queen (reproductives) have a long life, spanning decades, and the queen remains highly fertile throughout her adult life. Workers, on the other hand, are short-lived and sterile. We studied this species of termites, using a combination of high throughput techniques such as transcriptomics, metabolomics and lipidomics, to identify mechanisms that allow reproductives to live orders of magnitude longer than workers, while maintaining high fertility. Aging associated genes were differentially expressed between the reproductives and the workers. For example, antioxidant genes were highly expressed, and the membrane lipids were less damaged by oxidative stress, in the queens relative to workers. Contrary to expectations, we found that several members of the insulin/insulin-like growth factor signaling (IIS) pathway were upregulated in the queens. Normally this would indicate increased diversion of metabolism towards energy storage. However, we did not find excessive fat storage in the queens; simple sugars dominate in their hemolymph and a large amount of resources is allocated to egg production. Our findings support the notion that aging results from a complex interplay of several processes. These processes should be studied simultaneously and not in isolation, for a better understanding of aging.
20.10.2021
Convergent loss of chemoreceptors across independent origins of slave-making in ants

Socially parasitic ants exploit the work force and social organisation of closely related ant species, and therefore no longer need to perform certain social tasks such as brood care and foraging. The loss of such behaviours is expected to be accompanied by loss of the underlying genes. We sequenced the genomes of eight ant species representing three independent origins of parasitism. Due to their importance in chemical communication and foraging, we investigated the evolution of chemoreceptors in parasites and their hosts. We found that parasites lost a striking 50% of their gustatory receptor repertoire compared to their hosts, perhaps reflecting the outsourcing of foraging tasks to host workers. We also found that parasites had fewer olfactory receptors than their hosts, with the same olfactory receptors being lost across multiple origins of parasitism. This represents a rare case of convergent molecular evolution at the level of individual genes, shedding light on the loss of important social traits during the transition to a parasitic lifestyle.
12.10.2021
Ancestral sequences of a large promiscuous enzyme family correspond to bridges in sequence space in a network representation

Evolutionary relationships of protein families can be characterized either by networks or by trees. Whereas trees allow for hierarchical grouping and reconstruction of the most likely ancestral sequences, networks lack a time axis but allow for thresholds of pairwise sequence identity to be chosen and, therefore, the clustering of family members with presumably more similar functions. Here, we use the large family of arylsulfatases and phosphonate monoester hydrolases to investigate similarities, strengths and weaknesses in tree and network representations. For varying thresholds of pairwise sequence identity, values of betweenness centrality and clustering coefficients were derived for nodes of the reconstructed ancestors to measure the propensity to act as a bridge in a network. Based on these properties, ancestral protein sequences emerge as bridges in protein sequence networks. Interestingly, many ancestral protein sequences appear close to extant sequences. Therefore, reconstructed ancestor sequences might also be interpreted as yet-to-be-identified homologues. The concept of ancestor reconstruction is compared to consensus sequences, too. It was found that hub sequences in a network, e.g. reconstructed ancestral sequences that are connected to many neighbouring sequences, share closer similarity with derived consensus sequences. Therefore, some reconstructed ancestor sequences can also be interpreted as consensus sequences.
30.03.2021
New research priorities in Münster

Die Deutsche Forschungsgemeinschaft (DFG) fördert zwei neue Schwerpunktprogramme (SSP) an der Westfälischen Wilhelms-Universität in Münster. Insgesamt fließen dafür in den nächsten drei Jahren 10 bis 14 Millionen Euro nach Münster. Gefördert werden Programme in der Biologie und der Chemie. Im Projekt „Die genomischen Grundlagen evolutionärer Innovationen (GEvol)“ unter der Leitung des Biologen Prof. Dr. Erich Bornberg-Bauer vom Institut für Evolution und Biodiversität wollen die Forscher die Mechanismen der genetischen Veränderungen einer großen Artengruppe entschlüsseln. „In dem Vorhaben untersuchen wir unter anderem die Prozesse, die den wichtigsten genomischen Veränderungen in der Evolution zugrunde liegen – beispielsweise Gewinn und Verlust von Sozialität oder Paarungssystemen, Verteidigung und Immunität, entwicklungsbiologischen und morphologischen Anpassungen“, sagt Erich Bornberg-Bauer. Read more
29.03.2021
Münster University receives two new research associations

The University of Münster is coordinating two new Priority Programmes funded by the German Research Foundation (DFG) with several million euros. The projects come from the fields of biology and chemistry and deal with innovative informatics technologies.Read more
12.03.2021
Structural and functional characterization of a putative de novo gene in Drosophila

Comparative genomic studies have repeatedly shown that new protein-coding genes can emerge de novo from noncoding DNA. Still unknown is how and when the structures of encoded de novo proteins emerge and evolve. Combining biochemical, genetic and evolutionary analyses, we elucidate the function and structure of goddard, a gene which appears to have evolved de novo at least 50 million years ago within the Drosophila genus. Previous studies found that goddard is required for male fertility. Here, we show that Goddard protein localizes to elongating sperm axonemes and that in its absence, elongated spermatids fail to undergo individualization. Combining modelling, NMR and circular dichroism (CD) data, we show that Goddard protein contains a large central α-helix, but is otherwise partially disordered. We find similar results for Goddard’s orthologs from divergent fly species and their reconstructed ancestral sequences. Accordingly, Goddard’s structure appears to have been maintained with only minor changes over millions of years.
12.03.2021
New proteins 'out of nothing'

Proteins are the key component in all modern forms of life. Haemoglobin, for example, transports the oxygen in our blood; photosynthesis proteins in the leaves of plants convert sunlight into energy; and fungal enzymes help us to brew beer and bake bread. Researchers have long been examining the question of how proteins mutate or come into existence in the course of millennia. That completely new proteins - and, with them, new properties - can emerge practically out of nothing, was inconceivable for decades, in line with what the Greek philosopher Parmenides said: "Nothing can emerge from nothing" (ex nihilo nihil fit). Working with colleagues from the USA and Australia, researchers from the University of Münster (Germany) have now reconstructed how evolution forms the structure and function of a newly emerged protein in flies. This protein is essential for male fertility. The results have been published in the journal "Nature Communications". Read more
19.02.2021
Marie Skłodowska-Curie Actions | Individual Fellowship awarded to Dr. Bertrand Fouks

How genomes evolve and drive novelty is a central question in biology. Some of the most puzzling genomic innovations, for example the development of placenta in mammals, are triggered by Transposable Elements (TEs). TEs are small genome fragments that can move and insert in other areas of the genome, which can create or impair gene functions. Organisms have adapted mechanisms to counteract the harmful effects of TEs, notably small RNAs (e.g. piwi-interacting RNA, piRNAs). Despite increasing knowledge on the effects of TEs on genome evolution and the apparition of novel traits, how and which TEs along with their interactions with piRNAs can promote novelty remain unclear. The project of Dr. Fouks will shed light on this issue by investigating how TEs and piRNAs evolved and interacted in cockroaches and termites alongside the evolution of their incredible biodiversity, with an emphasis on sociality and wood feeding. Dr. Fouks will generate several high-resolution genomes and transcriptomes from cockroach and termite species to locate and categorize TEs and piRNAs., This will allow him to unravel their role in the adaptation of cockroaches and termites to different social levels and diets.
24.07.2020
Humboldt Fellowship for Dr. Anna Grandchamp

Since several years it is known that new proteins not only arise via gene duplication and variation of the duplicates but also de novo, i.e. from previously non-coding DNA. An important first step in the creation of these de novo genes is that some of the zillions of randomly generated transcripts have some, though very weak, inherent function or are at least not toxic to the cell and are not quickly lost again. In her project, Dr. Grandchamp will investigate how often new random transcripts are created, by which mechanisms they are created and what the initial function of the new proteins might be.She plans to use in-bred lines of fly populations collected from all over Europe as well as of closely related fly species and map their transcriptomes onto the newly sequenced genomes to precisely characterise the creation and loss of de novo genes.
22.10.2019
Researchers gain new insights into the evolution of proteins

How do bacteria manage to adapt to synthetic environmental toxins and to even develop strategies for using a pesticide agent as food within less than 70 years? This is what scientists at Münster University have investigated. They found out how mutations led to biochemical changes that now enable an enzyme to cleave a pesticide. The study was published in "Nature Chemical Biology". Read more
11.09.2018
Bioinformaticians examine new genes the moment they are born

Accumulating evidence suggests that new genes can arise spontaneously from previously non-coding DNA instead of through the gradual mutation of established genes. Bioinformaticians at the University of Münster are now, for the first time, studying the earliest stages in the emergence of such “genes out of thin air”, also known as de novo genes. Read more
26.04.2018
"The Elusive Calculus of Insect Altruism

In 1964, the evolutionary biologist William D. Hamilton seemingly explained one of the greatest paradoxes in biology with a simple mathematical equation. Even Charles Darwin had called the problem his “one special difficulty” a century earlier in On the Origin of Species, writing that it made him doubt his own theory. The paradox in question is the altruistic behavior exhibited most famously by social insects. Ants, termites, and some bees and wasps live in highly organized colonies in which most individuals are sterile or forgo reproduction, instead serving the select few who do lay eggs. Yet such behavior seemed to clearly violate the concept of natural selection and survival of the fittest, if “fittest” means the individual with the greatest reproductive success. The insects’ compulsory altruism—a form of extreme social behavior called eusociality—made little sense. Read more
06.04.2018
"Human Frontier Science Program" funds two projects involving Münster University researchers

In the selection phase for 2018, the prestigious “Program Grant” research award given by the international “Human Frontier Science Program” goes to two members of the Department of Biology at the University of Münster – to bioinformatics specialist Prof. Erich Bornberg-Bauer and cell biologist Prof. Karin Busch. Read more
22.03.2018
Consider the Cockroach

On the fringes of urban life, some creatures thrive more than others. The brown rat, the coyote, the Canada goose, the Northern raccoon and many other species have all expanded their ranges and populations because they are well-adapted to scavenge or hunt in the shadows of human civilization. But perhaps none have been as successful as the cockroach. Read more
07.02.2018
The social evolution of termites
One phenomenon that already fascinated Charles Darwin is the evolution of huge, complex insect societies from solitary ancestors. This was the case with termites and ants, which have the same eusocial lifestyle. This has distinctive features such as the creation of castes, including for instance a complex system of division of labor among workers and soldiers. A team headed by evolutionary biologist Prof. Dr. Judith Korb from the University of Freiburg, bioinformatician Prof. Dr. Erich Bornberg-Bauer, evolutionary biologist Dr. Mark Harrison and evolutionary biologist Dr. Evelien Jongepier from the Westphalian Wilhelms University in Münster has now compared the molecular basis for the evolution of the eusocial lifestyle. Read more
05.02.2018
Scientists investigate the molecular basis of social evolution in Termites

Researchers from the group of bioinformatician Prof Erich Bornberg-Bauer from the Institute for Evolution and Biodiversity at the WWU have now, for the first time, compared the molecular basis for the evolution of eusociality within termites and ants. Read more
01.02.2016
Land plant became key marine species
The genome of eelgrass (Zostera marina) has now been unveiled. It turns out that the plant, once land-living but now only found in the marine environment, has lost the genes required to survive out of the water. Scientists from the University of Gothenburg participated in the research study, the results of which are published in the scientific journal Nature. Eelgrass belongs to a group of flowering plants that have adapted to a life in water. As such, it is a suitable candidate for studies of adaptation and evolution. 'Since flowering plants have emerged and developed on land, eelgrass can be expected to share many genetic features with many land plants. Studying differences between them can tell us how eelgrass has adapted to a marine environment,' says Mats Töpel, researcher at the Department of Marine Sciences, University of Gothenburg, who participated in the sequencing of the eelgrass genome. Töpel is part of an international research collaboration involving 35 research teams. As a result of their efforts, the eelgrass genome has now been published in Nature. Read more
31.08.2015
A Surprise Source of Life's Code

Genes, like people, have families — lineages that stretch back through time, all the way to a founding member. That ancestor multiplied and spread, morphing a bit with each new iteration. For most of the last 40 years, scientists thought that this was the primary way new genes were born — they simply arose from copie s of existing genes. The old version went on doing its job, and the new copy became free to evolve novel functions. Read more
20.05.2014
Tracking colony-building insects

Scientists have long been doing research into how the complicated system of living together in insect colonies functions. An international group of researchers – including scientists from Münster University – have now sequenced and analysed the genome of one type of termite. This means that they have now been able to compare the termites' DNA with that of ants and colony-building bees. The study has been published in "Nature Communications". Read more
09.04.2013
Prestigious award for bioinformatician

Prof. Erich Bornberg-Bauer from the Institute for Evolution and Biodiversity at Münster University has received a prestigious Program Grant from the international "Human Frontier Science Program" (HFSP). With this grant the HFSP supports outstanding scientists from various countries who jointly work on innovative research topics. Read more
12.07.2012
Enigma of Twisted-Wing Parasites resolved

Scientists have long been mystified by the insect group called twisted-wing parasite. It includes over 500 species and although it has been known for almost 200 years, till now it could not be linked to any super-ordinate group of insects. A team of scientists have, for the first time ever, sequenced the entire genetic code – the genome – of a twisted-wing parasite, thus enabling the scientists to classify these insects as a sister group of the beetle. Read more
11.02.2011
Secrets of fungus-growing

An international consortium of researchers, including Prof. Erich Bornberg-Bauer from Münster University's Institute of Evolution and Biodiversity, has been able top demonstrate for the first time that the highly specialized lifestyle of leafcutter ants has been deposited in the insects' genetic make-up. For example, they are missing certain genes which are otherwise necessary for digestion. Read more
18.01.2010
Bacteria change sperm of insects

Es ist ein recht gerissener Trick im Naturreich: Ein Bakterium sorgt dafür, dass sich sein Wirt erfolgreich vermehrt, indem er das Erbgut der Spermien verändert. Darin hatten Forscher überraschenderweise Teile des Bakterien-Genoms gefunden. Mit einem raffinierten genetischen Trick sichert das Bakterium Wolbachia sein Überleben in Insekten: Der Parasit manipuliert die Fortpflanzung seiner Wirte in großem Stil. Auf die Schliche gekommen ist dem Bakterium eine internationale Forschergruppe, die das Erbgut von Erzwespen entziffert und darin ein Gen des Schmarotzers entdeckt hat. Read more