P3: Evolutionary Origin, Fixation and Functions of de Novo Protein Coding Genes
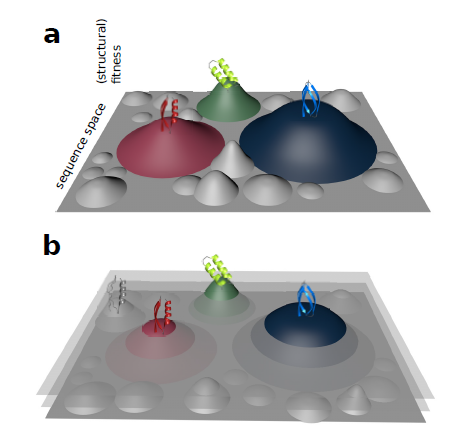
With every new genome sequenced a couple of hundred proposed genes remain ''orphans'' because computational methods could not assign any orthologs, even to closely related and well annotated species. Presumably many of these (lineage-specific) genes are transcribed, sometimes translated and proteins functional and adaptive, at least under some (possibly unknown) conditions. De novo emergence is not only against current believe that most novel genes emerge from old ones, it is also difficult to reconcile with a biophysical perspective because novel reading frames emerging from previously non-coding matter must be considered extremely unlikely: they would most likely be disordered, aggregate and thus be deleterious or, at least be purged for purely energetic reasons. So, where do new coding genes actually come from, how do they function and how is their -- potentially detrimental -- expression regulated?
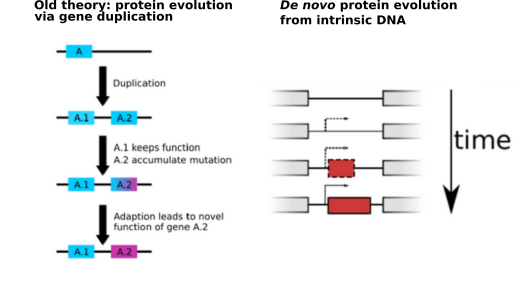
We ask where novel protein coding genes come from and how genomic novelties and rearrangements trigger adaptation and spur developmental transitions.
Using comparative genomics and biophysical analyses (computational and experimental) we test their properties and functions. We found that most genetic
novelty comes from novel domains but also many completely new reading frames emerge, e.g. across the insect tree, with an estimated frequency of 500
new genes in the wake of each speciation event. This former process has been termed ''grow slow and moult'' because some novel domains later lose their
initially stabilising parent protein and become independent and amenable for further rearrangements. We concentrate on some major transitions which
happened during the development of extant life forms: signalling across multicellular organisms, placentation in mammals, the emergence of holometabolism
in insects and the onset and reversal of ageing.
Furthermore, to catch novel genes "in the act" of emergence, we investigated genomes
not only between species but also from populations and, as an outgroup, their closely related sister species. We determine, using gene
and domain prediction programmes, novel ORFs, their expression (RNAseq) and, if necessary, confirm them e.g. with (long-read and primer walking)
PCR and qPCR. We are currently screening several systems (populations of fish, mice, flies, and human) to achieve a good genomic coverage for
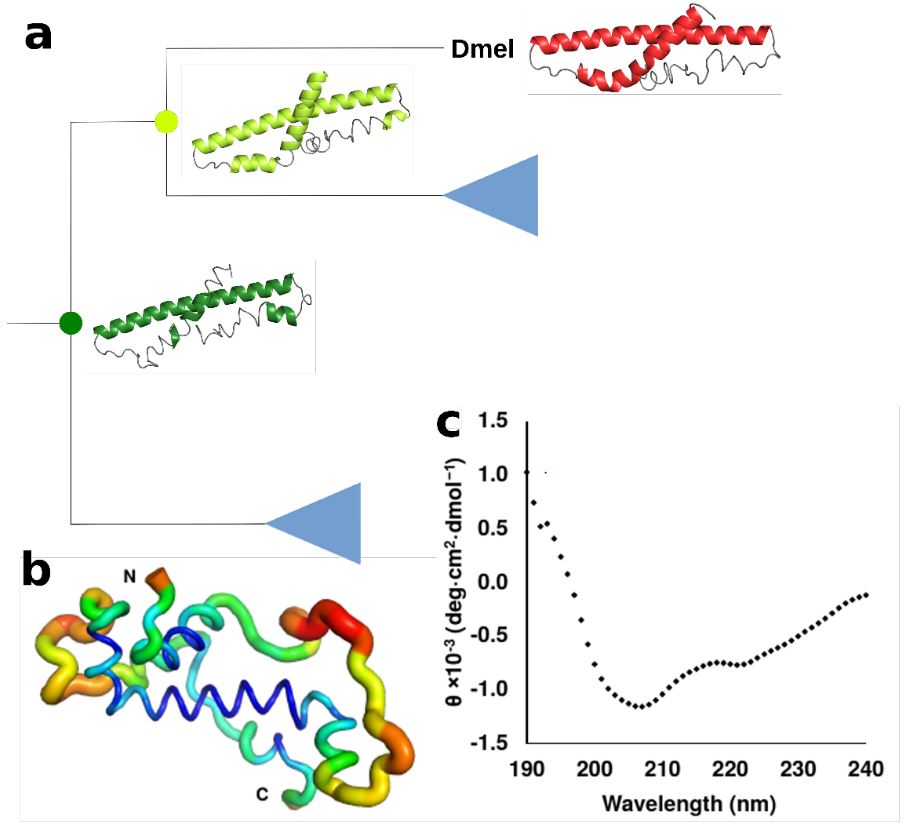
detecting possible recent emergence and reconstruct ancestral sequences which can then be tested for their genetic origin and investigate their
structural and biophysical properties with the help of TSA, CD, NMR, and phage display experiments. Additonally, we aim to examine the behavior
of the predicted ancestoral de novo gene compared to the existing one in vitro Drosophila experiments.
People: Andreas Lange, Anna Grandchamp, Bharat Ravi, Brennen Heames, Lars Eicholt, Margaux Aubel
Collaborations
- Geoffrey Findlay (Holy Cross, MA)
- Klara Hlouchova Prague
- Ylva Ivarsson (Uppsala)
- Florian Hollfelder (Cambridge)
- Colin Jackson (Canberra)
Funding: Leibniz Gemeinschaft (2013 -- 2016); Horizon 2020 Research and Innovation Framework Programme No. 722610 (2017 -- 2021); Volkswagen Stiftung (2021 -- 2026)
Related Publications
- Eicholt, LA, Aubel, M, Berk, K, Bornberg-Bauer, E, Lange, A. Heterologous expression of naturally evolved putative de novo proteins with chaperones. Protein Science. 2022; 31( 8):e4371. Online Access
- Lange, A, Patel, PH, Heames, B, Damry, AM, Saenger, T, Jackson, CJ, Findlay, GD, Bornberg-Bauer E; Structural and functional characterization of a putative de novo evolved gene essential for male fertility in Drosophila, Nat Comm, 2021:12(1667), Online Access
- Bornberg-Bauer E, Hlouchova, K, Lange A; Structure and Function of Naturally Evolved de novo Proteins, Curr Opn Struct Biol, 2021, Online Access
- Bornberg-Bauer, E. and Heames, B.; Becoming a de novo gene; Nature Ecology & Evolution, 2019 Online Access
- Schmitz JF, Ullrich K, Bornberg-Bauer E; Incipient de novo genes can evolve from "frozen accidents" which escaped rapid transcript turnover; Nature Ecology and Evolution, 2018 Online Access
- Klasberg, S, Bitard-Feildel, T, Callebaut, I and Bornberg-Bauer, E; Origins and Structural Properties of Novel and De Novo Protein Domains During Insect Evolution.; FEBS Journal, 2018 Online Access
- Schmitz, JF and Bornberg-Bauer E; Fact or fiction: Updates on how protein coding genes might emerge de novo from previously non-coding DNA; F 1000Research, 2017 Online Access
- Gubala et al. The goddard and saturn genes are essential for Drosophila male fertility and may have arisen de novo; Molecular Biology and Evolution, 2017 Online Access
- E Bornberg-Bauer et al. Emergence of de novo proteins from ''dark genomic matter'' by ''grow slow and moult''; Transactions of the Biochemical Society; 2015. Online Access
- T Bitard-Feildel et al. Detection of Orphan Domains in Drosophila using Hydrophobic clustering analysis; Biochimie, 2015 Online Access
- L. Wissler et al. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 2013 Online Access
- P. Feulner et al. Genome-wide patterns of standing genetic variation in a natural marine population of three-spined sticklebacks. Molecular Ecology 2012 Online Access
- F. Chain et al., Extensive copy-number variation of young genes across stickleback populations. PLoS Genet, 2014. Online Access
- E Bornberg-Bauer et al. How do new proteins arise?, Curr Opn Struct Biol, 2010. Online Access
Techniques employed: Computational: comparative genomics, differential GO analysis, biophysical predictions
(disorder, secondary structure, hydrophobic clusters), ancestral reconstruction and phylogenies, mutational effects on stability
(FoldX, Rosetta); experimental: deep sequencing, qPCR; antibody staining; cloning, (over-)expression, purification; SDS page
expression quantification; E.coli autodisplay (Jose); in-situ hybridisation; CD; stability measures; in-cell NMR (Selenko);
pull down assays (Ivarsson); in vitro expression of ancestral de novo genes (Findlay)