P2: Algorithms and Evolutionary Dynamics of Modular Protein Evolution
Modularity is a hallmark of molecular evolution, whether considering gene regulation, the components of metabolic pathways or signalling cascades,
the ability to reuse autonomous modules in different molecular contexts can expedite evolutionary innovation. Over times scales of several 100 MY,
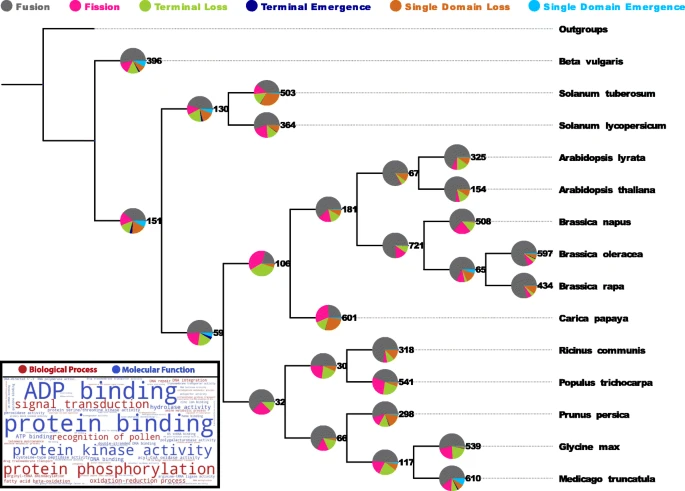
the evolution of protein coding genes is dominated by the modular rearrangements of protein domains, their evolutionary, structural and functional units.
While a small core of arrangements is universal, a large fraction of multi-domain arrangements is species specific and has been created recently via gene duplication,
fusion and terminal loss of domains. Surprisingly, thousands of domains are completely (i.e. all copies within a genome) lost from genomes along every lineage in a stochastic manner,
i.e. at a fairly constant rate of ca. four domains per million years.
Novel domains are rarely fixed and arise either as their own genes or terminally, by extension of existing reading frames.
Novel domains are under strong selection pressure and confer a strong fitness value as they rapidly attain high copy numbers
within the genomes and are involved in biotic defence, reproduction and development.
Using cross-species genomic comparisons
and population genomics we investigate the genetic mechanisms and biophysical constraints of domain emergence and develop algorithms
for rapid screening of many genomes, understanding their phylogenies and comparing, aligning and clustering sequences.
This is possible because of complexity reduction (sequences can be characterised as linear arrangements of 5-6 domains drawn
from an alphabet of several thousand characters as opposed to ca. 500 amino acids drawn from an alphabet of 20) and the maintenance
of linear order and the high reliability of HMMs which characterise the domains.
We test the potential of increased evolvability by
rearranged domains experimentally in biochemical pathways, for the defence against pathogens and during developmental processes.
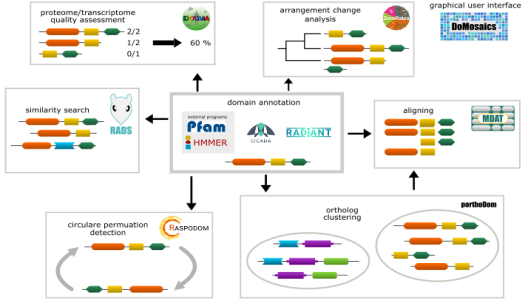
The software we develop is available here: DomainWorld
People: Carsten Kemena, Steffen Klasberg, Elias Dohmen, Baki Coban
Collaborations
Funding: Volkswagen Foundation (2 x, 2009 -- 2013); DFG (2009 -- 2013).
Related Publications
- E. Bornberg-Bauer Computational Approaches to Identify Leucine Zippers. Nucleic Acids Res. 1998 Online Access
- J. Weiner, G. Thomas and E. Bornberg-Bauer Rapid Motif-Based Prediction of Circular Permutations in Multi-Domain Proteins. Bioinformatics 2005 Online Access
- J. Weiner and E. Bornberg-Bauer Evolution of Circular Permutations in Multi-Domain Proteins. Molecular Biology and Evolution 2006 Online Access
- Andrew D. Moore and Erich Bornberg-Bauer The Dynamics and Evolutionary Potential of Domain Loss and Emergence. Molecular Biology and Evolution 2012 Online Access
- E. Bornberg-Bauer and M.Mar Alba; Dynamics and Adaptive Benefits of Modular Protein Evolution, Curr Opn Struct Biol 2013. Online Access
- A. Schüler and E. Bornberg-Bauer, Evolution of Protein Domain Repeats in Metazoa, MolBiolEvol, 2016 Online Access
- E. Dohmen, L.P.M. Kremer, E. Bornberg-Bauer and C. Kemena, DOGMA: Domain-based transcriptome and proteome quality assessment, Bioinformatics, 2016 Online Access
- S. Klasberg, T. Bitard-Feildel, I. Callebaut, and E. Bornberg-Bauer, Origins and Structural Properties of Novel and De Novo Protein Domains During Insect Evolution. FEBS Journal, 2018 Online Access
- GWC Thomas, E Dohmen et al. The Genomic Basis of Arthropod Diversity, Genome Biology, 2020 Online Access
- E. Dohmen, S. Klasberg, E. Bornberg-Bauer, S. Perrey and C. Kemena, The modular nature of protein evolution: Domain rearrangement rates across eukaryotic life, BMC Evolutionary Biology, 2020 Online Access
Techniques employed: Hidden Markov Models, dynamic programming, design and implementation of algorithms, databases and interactive graphical interfaces; DnDs ratio tests (PAML)